Installation
CoMPaseD is written in Python 3 and has been tested on Windows (≥ Windows 10) and Linux machines. Installation and program start should be done via the command prompt, while configuring and running CoMPaseD analyses is possible via the command prompt or the graphical interface.
Dependencies
Crux Toolkit
Binaries for the crux mass spectrometry analysis toolkit are available via the official crux webpage. Download the binaries suitable for your operating system (Windows/Linux) (version ≥ 3.2).
Python
To run CoMPaseD python (versions 3.5–3.12 tested) is required.
Note
While newer python versions may be compatible, some dependencies may not be immediately available for the latest releases. It is recommended to use a tested version to ensure full functionality.
Install python using:
Download Python for your Windows distribution.
Install Python by executing the downloaded file and following the instructions.
Ubuntu/Debian:
sudo apt update sudo apt install -y python3 python3-pip
Fedora
sudo dnf install -y python3 python3-pip
Arch Linux
sudo pacman -Syu python python-pip
Python packages
The Python packages with suitable version numbers are listed in the provided requirements.txt.
requirements.txt
Package name |
Version |
|---|---|
bio |
≥1.5.9 |
biopython |
≥1.81 |
colorama |
≥0.4 |
keras |
≥2.13.1 |
matplotlib |
=3.7.2 |
numpy |
≥1.22 |
pandas |
≥2.0 |
PyQt6-Qt6 |
≥6.5 |
PyQt6-sip |
≥13.5 |
PyQt6 |
≥6.5 |
python-dateutil |
|
tensorflow-estimator |
|
tensorflow-intel |
|
tensorflow-io-gcs-filesystem |
|
tensorflow |
≥2.12.0 |
Tip
We recommend using a virtual environment and pip to install these dependencies.
Install the Python requirements using pip and requirements.txt:
Set Installation Directory
Open a command-line window and navigate to the directory where you want to install CoMPaseD:
mkdir C:\Programs\CoMPaseD cd C:\Programs\CoMPaseD
Clone the Repository
Copy the CoMPaseD repository into the chosen directory:
git clone https://github.com/MicrobialProteomics/CoMPaseD.git
Tip
The Git command may not be available by default in many Windows installations. You may need to install Git in advance.
Create and Activate a Virtual Environment
Set up a virtual environment to manage dependencies:
C:\Programs\python\python.exe -m venv venv C:\Programs\CoMPaseD\venv\Scripts\activate.bat
Install Dependencies
Install the required Python packages:
python -m pip install -r requirements.txt
Run CoMPaseD
Launch the graphical interface:
CoMPaseD_gui.py
Configure Crux Path
Set the path to the Crux toolkit within the GUI and save the configuration. CoMPaseD is now ready to use.
Set Installation Directory
Open a command-line window and navigate to the directory where you want to install CoMPaseD:
mkdir -p ~/Programs/CoMPaseD cd ~/Programs/CoMPaseD
Clone the Repository
Copy the CoMPaseD repository into the chosen directory:
git clone https://github.com/MicrobialProteomics/CoMPaseD.git
Tip
If you do not have
gitinstalled. Install it usingsudo apt install gitor your prefered package manager.Create and Activate a Virtual Environment
Set up a virtual environment to manage dependencies:
python -m venv venv source venv/bin/activate
Install Dependencies
Install the required Python packages:
python -m pip install -r requirements.txt
Run CoMPaseD
Launch the graphical interface:
CoMPaseD_gui.py
Configure Crux Path
Set the path to the Crux toolkit within the GUI and save the configuration. CoMPaseD is now ready to use.
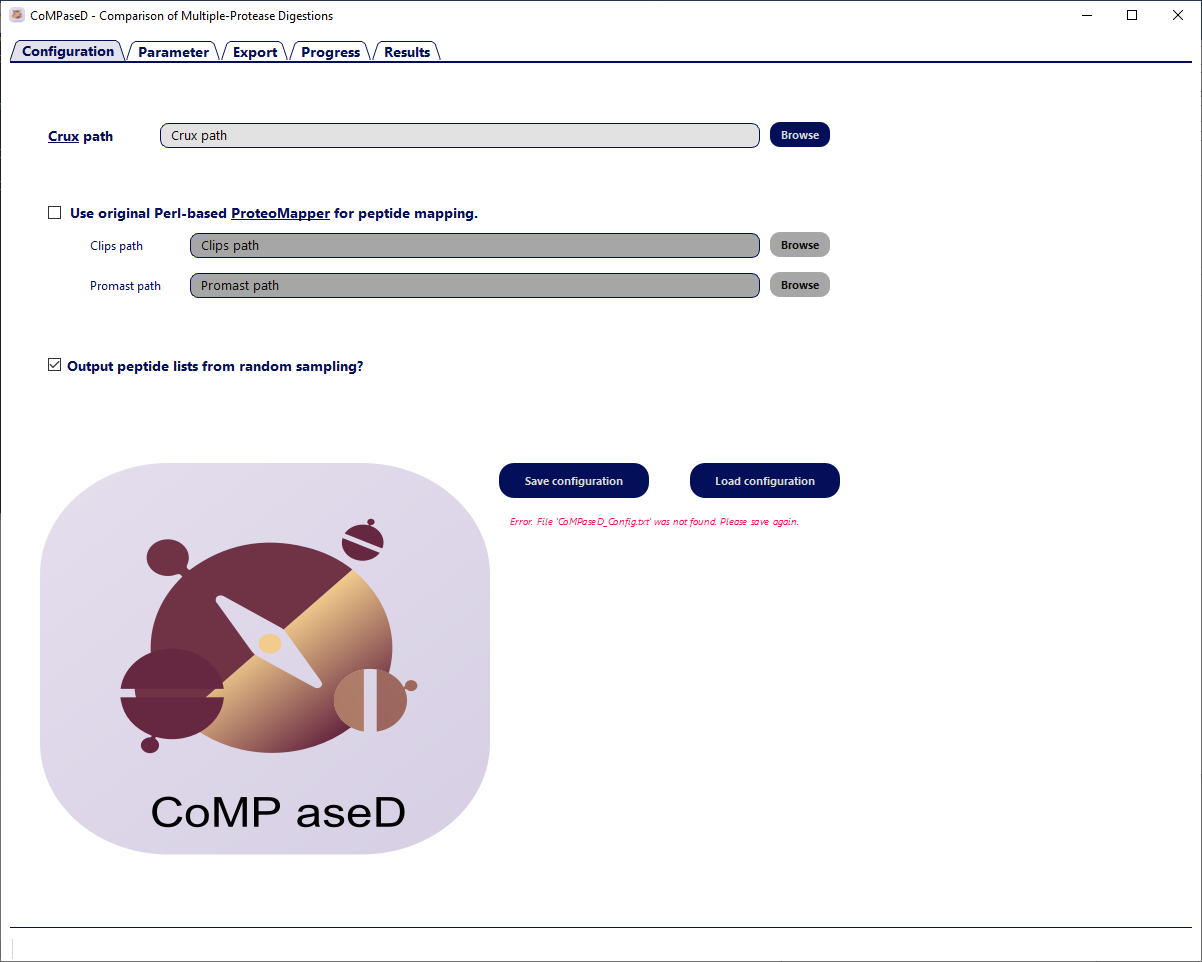
Configuration tab upon first start of CoMPaseD. The red error message will disapear when the path to the crux executable is properly set and the configuration is saved.
You can now start CoMPaseD at any time by activating the virtual environment and running:
CoMPaseD_gui.py(Graphical Interface)CoMPaseD_cli.py(Command-Line Interface)